Terminology
Gene Fusions
Gene fusions are a complex class of genomic variation that may be characterized by a broad range of relevant attributes with varying specificity.
A gene fusion is the joining of two or more genes resulting in a chimeric transcript and/or a novel interaction between a rearranged regulatory element with the expressed product of a partner gene (a regulatory fusion).
Genetic variations involving Genomic Rearrangements within the same gene (e.g. internal tandem duplications), and transcript alterations due to splice site variants, have similar structural properties (i.e. novel adjoining transcript segments) but are not considered gene fusions as they do not involve multiple genes.
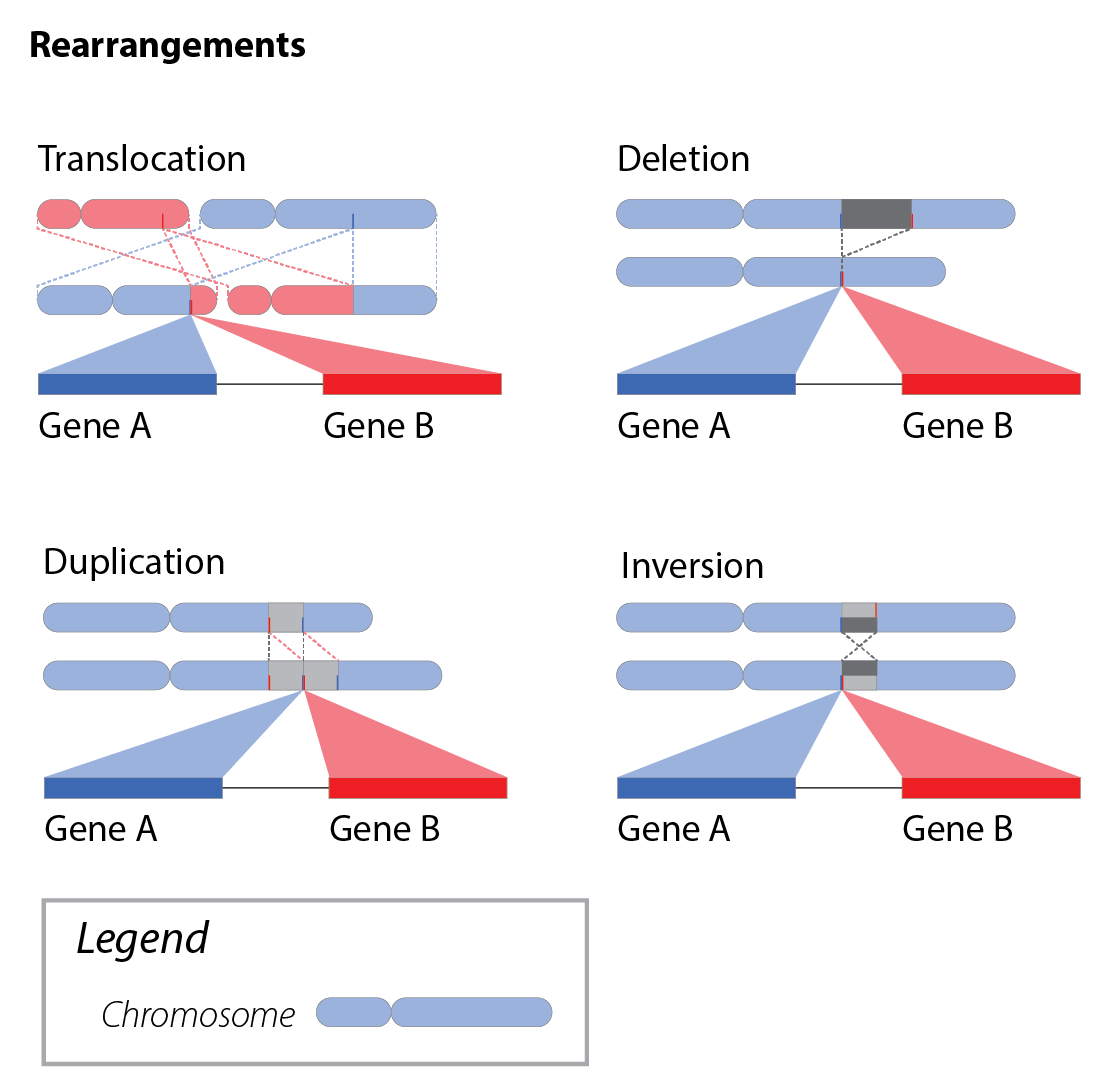
Importantly, gene fusions are also distinct from Genomic Rearrangements, though these concepts are often conflated due to the role of genomic rearrangements in creating gene fusions.
The two primary classes of gene fusions–Chimeric Transcript Fusions and Regulatory Fusions–are not mutually exclusive classes, as some fusions (such as promoter-swap fusions) may be defined either in the context of their regulatory elements or by their chimeric gene product.
Genes that are rearranged resulting in loss of an expressed product do not meet these definitions, and should not be described as gene fusions.
Chimeric Transcript Fusions
Chimeric transcript fusions are often driven by genomic rearrangements involving two gene loci, resulting in the concatenation of segments from each into a single transcript. This class of fusions is exemplified by well-known clinically-relevant gene fusions such as BCR::ABL1.
A chimeric transcript is a transcript (RNA molecule) composed of transcript segments from two or more genes.
Other biologically-relevant chimeric transcript fusions may be driven by RNA processing mechanisms in lieu of genomic rearrangements. One such mechanism is read-through transcription (e.g. CTSD-IFITM10), also known as tandem chimerism, where consecutive genes on a chromosome strand are transcribed as a single molecule prior to splicing [Akiva P, et al.]. Another mechanism is trans-splicing (e.g. trans-spliced JAZF1::JJAZ1 [Li H, et al.]), where two distinct transcripts are spliced together during processing.
Note: the special case of read-through fusions
Descriptions of read-through transcripts typically involve genes that are adjacent to one another in a reference genome assembly. The duality of the component genes encoding both independent and joint transcripts has resulted in authorities such as HGNC and CCDS creating read-through gene concepts, following a GENE1-GENE2 naming convention. However, read-through transcripts generated from genes brought into proximity by a rearrangement should be annotated with a double-colon instead of a hyphen.
In practice, it is rare to report on read-through events, as these events are common and typically have little known biological relevance [Mudge J].
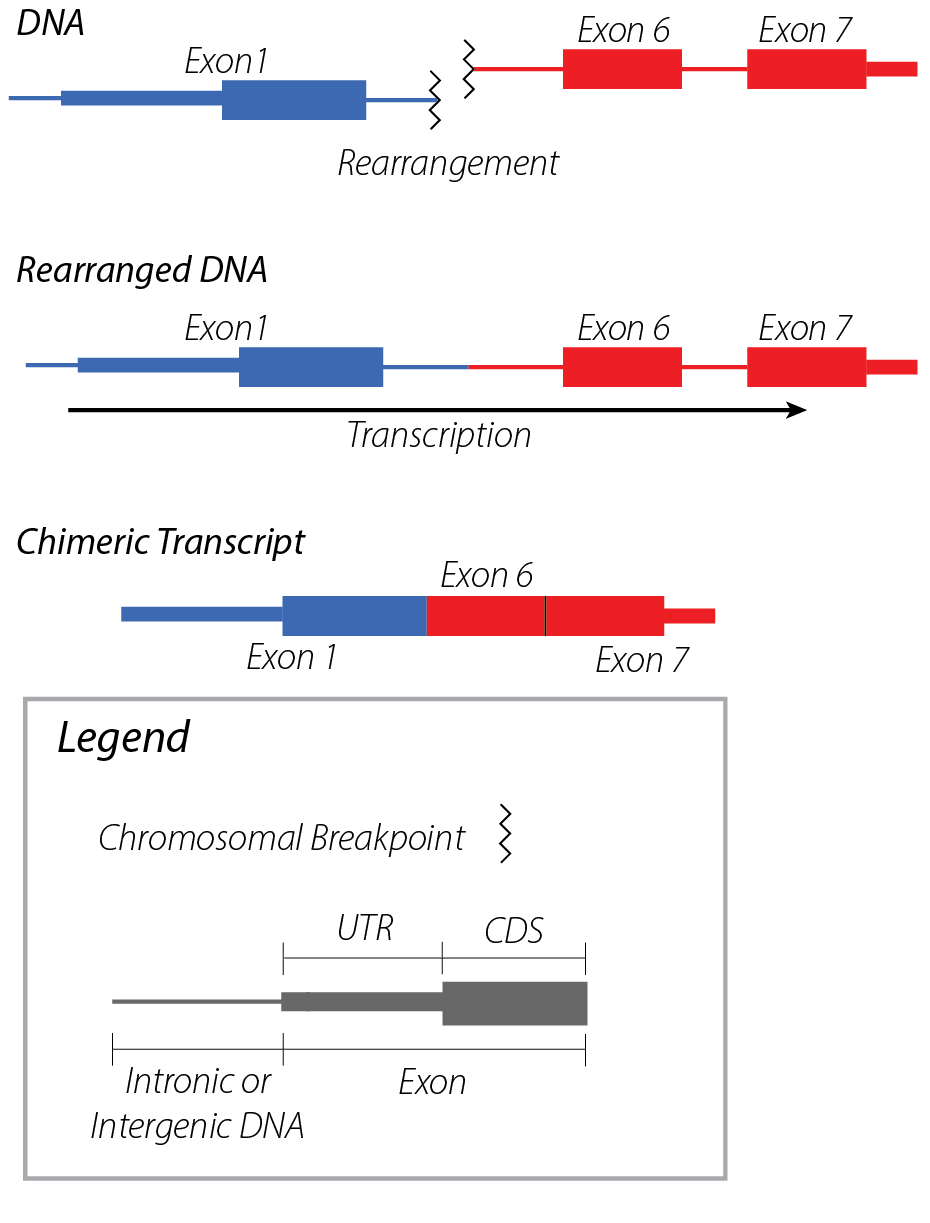
Gene fusions typically result in chimeric transcripts between two genes, which (for coding transcripts) often result in novel protein sequences. These chimeric transcripts are often caused by DNA rearrangements that bring the DNA elements contributing to a gene fusion into close proximity with one another.
Regulatory Fusions
Regulatory fusions are characterized by the rearrangement of regulatory elements from one gene near a second gene, typically resulting in the increased gene product expression of the second gene. This class of gene fusions should be described using the Regulatory Nomenclature, and includes promoter-swapping gene fusions such as reg_p@TMPRSS2::ERG, as well as enhancer-driven gene fusions such as reg_e@GATA2::EVI1.
A regulatory fusion is the novel interaction of a regulatory element brought into proximity of a partner gene by a genomic rearrangement, modulating gene product expression of the partner gene.
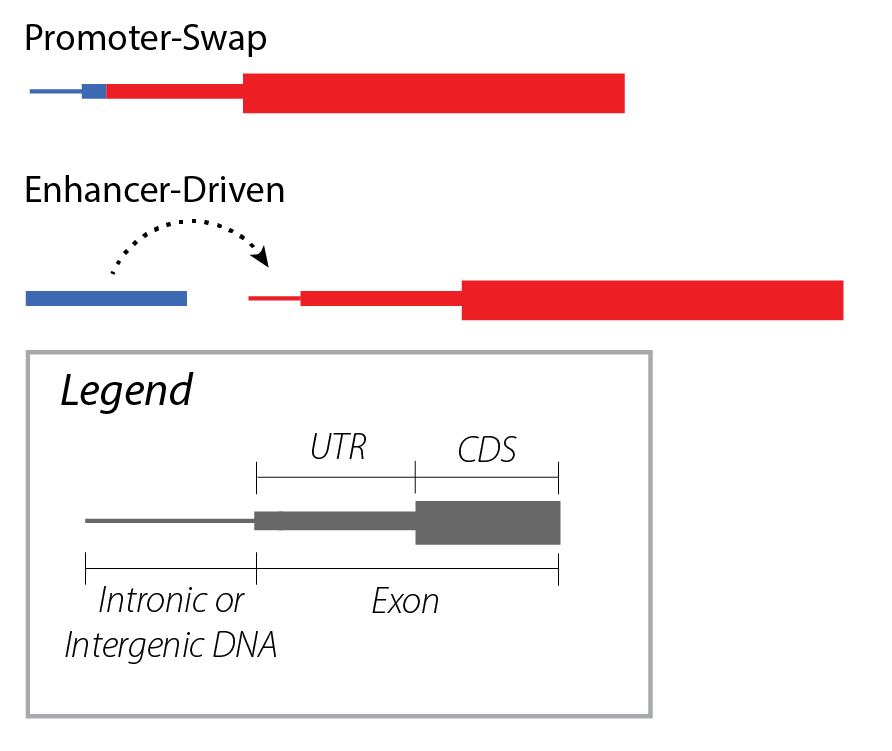
Gene fusions may be regulatory in nature, where a rearranged promoter or nearby enhancer element drives overexpression of the partner gene.
Gene Fusion Contexts
Determining the salient elements for a gene fusion is dependent upon the context in which the gene fusion is being described, whether it describes an assayed fusion event from a sample (Assayed Gene Fusions) or an aggregate context described in biomedical literature or knowledgebases (Categorical Gene Fusions). This specification provide recommendations for characterizing gene fusions in each context.
Assayed Gene Fusions
Assayed gene fusions from biological specimens are directly detected using RNA-based gene fusion assays, or alternatively may be inferred from genomic rearrangements detected by whole genome sequencing or cytogenomic assays in the context of informative phenotypic biomarkers. For example, an EWSR1 fusion is often inferred by breakapart FISH assay when a neoplasm is diagnosed or suspected to be Ewing sarcoma/primitive neuroectodermal tumor by immunohistochemical and/or morphological analysis.
Categorical Gene Fusions
In contrast, categorical gene fusions are generalized concepts representing a class of fusions by their shared attributes, such as retained or lost regulatory elements and/or functional domains, and are typically curated from the biomedical literature for use in genomic knowledgebases. Example categorical gene fusions include:
EWSR1 as a known 5’ gene fusion partner that joins one of many putative 3’ partner genes
ALK as a 3’ gene fusion partner with a retained kinase domain, which joins one of many putative 5’ partner genes
The class of BCR::ABL1 fusions involving multiple possible junctions between exons from the constituent BCR and ABL1 transcripts